GC IMAGE - GC Image GCxGC Edition - User Guide 2024R3
New Features and Improvements in Version 2020
GC Image GCxGC Edition 2020 has many new features and improvements. See
the GC Image GCxGC Edition User Guide for full
documentation.
What's New in Version 2020 Release 3 (June 2021)
Version 2020R3 has the following improvements for subtracting
multiple background spectra from a chromatogram:
- Subtract
Spectrum from Image has improved handling of negative
normalization ratios. Negative normalization ratios are usually
caused by previous subtraction operations. By skipping subtraction on
these data points, the user can subtract multiple spectra of
different background interferences from a chromatogram more
conveniently.
- The MS View > Copy to
Clipboard menu now has new options for copying spectra with absolute
intensities, making it easier to specify a spectrum for the Subtract
Spectrum from Image step when editing
a method.
Version 2020R3 has new improvements for using a
project as a workspace, including:
- Register, Review, and Investigate Images actions
under the Project > Run
menu now support all images inside a project without requiring
associated runs.
- Summary, Filter, and Comparison reports under the Project > Reports menu now support all
images inside a project without requiring associated runs.
- When saving a new image within an instance of the Image
program launched from a project, the save destination will now
default to the project's Images folder.
- The Run Details and
Images Details panes now
display more complete source file path information for more
convenient data validation.
Version 2020R3 adds further improvements for batch processing for the
Command Line interface:
- The Command Line interface now supports processing images
with a method using the new '
-method
[Path to method file]' option. The user no longer needs to copy and
edit a method in a command file.
- Using the command line has been simplified with the new Cli.bat interface.
The new interface supports relative paths for arguments and can be
called from any file location. Calling the interface no longer
requires the use of output redirection operators.
- The Command Line interface now exits with an error
message if provided an invalid method or command file path instead of
silently skipping processing steps.
Additionally, the Save Summary Report, Export Image as
CSV, and Export Image as Text Format steps of a
method now support specifying the
destination folder within the method editor, which makes it easier to
use a method with the Command Line interface.
Version 2020R3 has other improved operations, including:
- The software now supports importing ChemStation data channels
from an Agilent OpenLab .dx raw data file.
- The default system settings can now be set within Investigator
through the new Tools > Configure Settings menu.
- Investigator
validates the source image file for file existence and software
association before opening externally to avoid unnecessary errors.
- Investigator
now shows the source file path of the selected image.
- The software now hides disabled image-specific options if no image
is open when Export Configuration.
What's New in Version 2020 Release 2 (April 2021)
Version 2020R2 introduces three new method
commands providing new options for subtracting background noise from
chromatograms during batch processing:
- Subtract an Image allows the user to subtract an image
(for example, a background image) from the image processed by the
method.
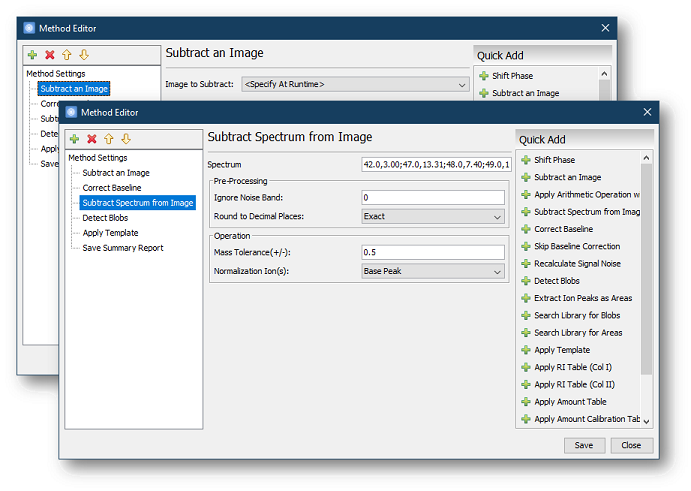
- Subtract Spectrum from Image allows the user to
subtract a specific spectrum from the image processed by the method.
For example, this command can be used to remove noise with a known
spectrum from the chromatogram.
- Apply Arithmetic Operation with an Image allows the
user to perform an arithmetic operation (addition, subtraction,
multiplication, division) between a specified image and the image
processed by the method. For example, addition of multiple
chromatograms can be used to obtain an average chromatogram.
In addition, Version 2020R2 has other related improvements, including:
- A new shortcut Processing
> Subtract an Image operation for subtracting an image from the
open image. The operation performs point-wise subtraction between the
two images.
- Processing
> Arithmetic Op > Browse now remembers recently used file
paths and includes buttons for selecting these paths. There are
buttons for selecting both the Recent Location (last used
during import) and the Image Location (the workspace images
folder).
Version 2020R2 has the following improvements for method usage, including:
- Method file parameter browsers now default to an appropriate
workspace folder, if available, making it easier to find the files
needed.
- The Use Method file browser from the Automated Actions tab of
the Import Options dialog now remembers
recently used file paths and includes buttons for selecting these
paths. There are buttons for selecting both the Recent
Location (last used during import) and the Method Location
(the workspace method folder).
- Unsupported method commands (such as those introduced in
future versions of the software) are now identified as such and
handled gracefully without causing errors.
Version 2020R2 adds a new data type for image export, MS for
Library Search. This new data type will generate each exported blob
or area spectrum using the library search settings set in Configure > Settings > Search
Library. The MS for Library Search data type is available from the Export
Image as Text Format method command as well as the following File > Export Image menu items:
Export Image as Text MS, Export Image as MzXML, Export Image as
Agilent CEF, and Export Image as Other Text Formats.
Additionally, Version 2020R2 improves the functionality of the OK
and View Spectrum button from the Blob
Properties dialog. The button now behaves the same as clicking on the
blob using the Context MS cursor mode, including listing both the peak
and blob MS spectra and selecting the active spectra based on the
library search settings.
What's New in Version 2020 Release 1.1 (September 2020)
Version 2020R1.1 has an updated batch search interface that is
fully compatible with the latest NIST 2020 libraries including the
updated RI library, and the new MS/MS libraries that double in coverage.
What's New in Version 2020 Release 1 (July 2020)
Version 2020R1 includes a new 64-bit framework that provides
improved performance and system stability for all operations when
processing data larger than 2GB. Due to the new framework, the
software no longer supports the Microsoft Windows Vista or older
operating systems.
Version 2020R1 introduces the Command Line interface
that allows processing a single raw data file or re-processing a
chromatographic image with a Method
or scripting. The interface can be used to create workflows that
integrate multiple tools or software for automating routine analysis
or complicated processing with a large number of data files.
Version 2020R1 has new improvements on generic
retention index calibration especially
for batch processing, including:
- A new Retention Index Method (.gcrm) file that allows
the user to export and import the RI table and settings together
either manually or through a Method;
- Default retention index calibration settings now can be
configured and exported through Configure
> Settings along with other settings;
- Import and Refresh an RI table now keeps current settings;
- A clear error message, instead of an empty table, is
displayed if a linked RI table file is empty or invalid.
In addition, the software now supports
- Base 10, in addition to natural log, for the log-linear
regression;
- Converting other retention properties of a blob to retention
indices with two new CLIC
functions, LRI1() and LRI2().
Version 2020R1 has two new filters for
Processing > Extract Ion
Peaks as Areas:
- New Min Qualifier Ion Count filter that allows the
user to specify the minimum number of apexing ions required;
- New CLIC Filter that allows the user to specify a
customized constraint using all properties computed for a targeted
ion apex peak, for example, retention times or a combination of peak
width and qualifier ions.
The operation now can be added to a Method,
and supported by batch processing with Project
or the Command Line interface.
In addition, the Context
MS mode has the following improvements to make it easier to examine
spectra deconvolved by the operation, including:
- Reference MS, where the deconvolved spectrum are
stored, is loaded to the MS View automatically;
- Only the primary spectra are set to be visible by default for
a less cluttered display;
- A spectrum can be set to Active and Visible separately to
allow fast toggling of the displayed spectra in MS Viewer.
Version 2020R1 has new improvements for
Investigator, including:
- Spectra view in the Compounds Perspective
now supports sending a selected spectrum to the NIST Library for
identification search.
- Compound Finder
now displays the results next to the main perspectives, which allows
cross examining the information of resulting compounds from the main
perspectives.
Version 2020R1 has other improved operations, including:
- Interactive
Blob Detection now has a new Apply and Save as Defaults
button that allows the user to save detection settings as the default
configuration. The button replaces the old Set-to-Configuration
option and question.
- Interactive
Blob Detection now sets only the Min threshold, instead of
both Min and Max thresholds, by default when SNR or other threshold-based
suggested properties are added.
- The software now supports smaller font sizes (i.e. 4 - 7)
than 8 for Text
annotation on small chromatogram images.
Contents
Next: Introduction
GC Image™ GCxGC Edition User Guide ©
2001–2021 by GC Image, LLC, and the University of Nebraska.